Institute for Structural and Chemical Biology
Facilities and technologies
The Institute provides access to world-class research facilities both within the Institute and the wider University.
Cryo-Electron Microscopy
Contact CryoEM@leicester.ac.uk for more information.
 FEI Titan Krios G3
FEI Titan Krios G3
- High-brightness X-FEG gun
- Three lens condenser system for parallel sample illumination
- 80 to 300 kV
- Constant Power lens design for optimal mode switching between LM-HM imaging and diffraction
- Autoloader for robotic sample handling and loading of up to 12 samples
- Falcon 3EC Camera
- Gatan K3 Camera with Bioquantum Energy Filter
- Volta Phase Plate for close to focus, high-contrast, data acquisition: Ideal for smaller (<200 kDa) macromolecular complexes
-
Thermo Fisher EPU: Software for automated single particle collection
- Falcon 3EC and K3 fully embedded
- Aberration free image-beam shift (AFIS) combined with fringe free illumination (FFI) yield up to 900 micrographs per hour on the K3.
- Thermo Fisher Batch Tomography: Software for automated tomogram collection
- Serial EM: Software for automated single particle and tomogram collection
 Philips CM200
Philips CM200
- High-brightness FEG gun
- 120 to 200 kV
- Side-entry Gatan 626 cryo-holder
- TVIPS F224HD 2k×2k CCD camera
 FEI Vitrobot
FEI Vitrobot
User friendly and reproducible, this plunger is ideal for new and experienced users. Double sided blotting and temperature and humidity control.
 Custom Manual Plunger
Custom Manual Plunger
This simple plunger offers high reproducibility with a small learning curve. The single sided blotting can be advantageous for some specimens, such as bacterial cells.
 Quorum GloQube
Quorum GloQube
A dual chamber glow-discharge machine for making grids hydrophilic with either air (negative charge) or a volatile additive such as pentalamine (positive charge).
Staff
- Professor John Schwabe - Director
- Dr Emma Hesketh - Facility Manager
- Dr TJ Ragan - Computational Facility Manager
- Dr Claudia Lancey - Experimental Officer
Nuclear Magnetic Resonance
 The Henry Wellcome Building is equipped with high field nuclear magnetic resonance (NMR) spectrometers which enable superior levels of sensitivity and resolution. The spectrometers posses cryoprobes that increase signal-to-noise ratio and allow NMR on samples of low concentration or faster acquisition of data. We are one of the few centres that combines both solution and solid state NMR to answer biological questions. Solution–state NMR allows the study of protein structure and dynamics at atomic detail and under physiological conditions, whereas solid-state NMR alleviates molecular weight limitations, allowing for the study of higher molecular weight complexes or of proteins embedded in lipid bilayers.
The Henry Wellcome Building is equipped with high field nuclear magnetic resonance (NMR) spectrometers which enable superior levels of sensitivity and resolution. The spectrometers posses cryoprobes that increase signal-to-noise ratio and allow NMR on samples of low concentration or faster acquisition of data. We are one of the few centres that combines both solution and solid state NMR to answer biological questions. Solution–state NMR allows the study of protein structure and dynamics at atomic detail and under physiological conditions, whereas solid-state NMR alleviates molecular weight limitations, allowing for the study of higher molecular weight complexes or of proteins embedded in lipid bilayers.
The NMR facilities include:
- Four NMR spectrometers (500 MHz, 2 X 600MHz, 800MHz)
- Including cryo-probes for increased signal-to-noise ratio
- One 600MHz dedicated to solid state NMR
For more information please contact Dr Fred Muskett at fwm1@leicester.ac.uk.
Scientific Computing
We have a dedicated computational expert, Dr TJ Ragan, to support and develop scientific computing and computational infrastructure. This allows us to be responsive to the needs of researchers to handle their data which is ever increasing in volume and complexity. The support provided includes:
- Scientific programming and training for researchers
- Developing and implementing new data analysis methods
- Assembling custom computational hardware for data processing
- High performance CPU- and GPU- based computing
The recent revolution in cryo-EM has resulted in the need to process very large (>2TB) data sets. A pair of GPU based computers tuned to process cryo-EM data have been built. These high performance boxes have reduced a typical run from days (on a supercomputer) to hours. Both computers are now in routine use for the analysis of medium- and high resolution electron microscopy datasets.
X-Ray Crystallography
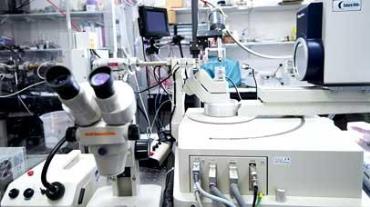 The X-ray facilities hosted in the Henry Wellcome Building provide users with the necessary infrastructure for researchers to prepare protein crystals, screen crystals and collect data in house. We also have regular access to the synchrotron facilities in the UK (Diamond Light Source, Oxford) and Europe (European Synchrotron Research Facility, Grenoble).
The X-ray facilities hosted in the Henry Wellcome Building provide users with the necessary infrastructure for researchers to prepare protein crystals, screen crystals and collect data in house. We also have regular access to the synchrotron facilities in the UK (Diamond Light Source, Oxford) and Europe (European Synchrotron Research Facility, Grenoble).
The X-ray crystallography facilities include:
- Rigaku MicroMax™ 007 HF rotating anode X-ray generator with multi-layer optics
- A HyPix-6000HE Hybrid Photon Counting (HPC) detector mounted on an AFC-10 K 4-circle goniometer
- custom single-crystal spectrophotometer
- TTP LabTech mosquito® liquid handling robot - Fast, high throughput crystallisation with minimal protein
- Douglas Instruments Oryx8 crystallisation robot - Crystal optimisation and microseeding capability
- 2 x Mosquito crystallisation robots- Nanolitre volume crystallisation
- Douglas Instruments Oryx4 crystallisation robot within an anaerobic chamber - Anaerobic crystallisation for oxygen sensitive systems
Biophysical Instrumentation Suite
The Biophysical Instrumentation Suite is run by Jaswir Basran. Please get in touch with Jaswir at wir@le.ac.uk for instrument booking, one-to-one training for inexperienced users, and help with experimental design and analysis.
Explore how to book instrumentation.
Instrumentation
Macromolecular interaction characterisation
- Octet QKE ForteBio Biolayer Interferometry (Sartorius)
- Stopped flow spectrometer (Applied Photophysics)
- Multiple Isothermal Titration Calorimetry instruments
Macromolecular quantification
- Horiba spectrofluorimeter (TCSP)
- PE lambda multicell spectrophotometer
- Cary Eclipse Fluorescence spectrometer
- Nanodrop 2000 spectrophotometer
Macromolecular characterisation
- Circular Dichroism
- SECMALS Dawn Heleos, Wyatt (JS)o
- Refeyn TwoMP Mass Photometer
- Nanosizer Dynamic Light Scattering analyser
Crystalisation robots
- Oryx crystallisation robots
- Anaerobic glove box 1 (Belle Technology) Oryx 4 (Douglas)
- Mosquito crystallisation robots
- Dragonfly crystallisation robot
Sample purification and preparation
- Multiple Aktas
- Peptide synthesiser
Steering committee
- Professor Peter Moody
- Professor Thomas Schalch
- Professor John Schwabe
- Dr Tennie Videler
Chemical Biology
 Within the Department of Chemistry researchers have access to first class facilities for both synthetic and physical chemistry research, these include:
Within the Department of Chemistry researchers have access to first class facilities for both synthetic and physical chemistry research, these include:
- NMR – Bruker DRX400, Bruker AV500, Bruker AV400
- X-ray Crystallography – Bruker APEX SMART 2000 Diffractometer
- Mass Spectrometry – Kratos Concept 1H, Micromass Quattro, Perkin-Elmer TurboMass, Applied Biosciences Voyager DE-STR, Waters Acquity XEVO Q ToF
- Scientific Glassblower - production of specialised glassware.
Chemical biology expertise at Leicester covers a broad range of synthetic capabilities and physical measurement methodologies, including:
Synthetic capabilities
- Novel small molecule libraries - Generation of bespoke small molecule libraries for emerging approaches such as the development of proteolysis targeting chimeras (PROTACs) and protein-protein interaction inhibitors
- Small and large-scale synthesis of peptides, peptoids, PNA and peptidomimetics. Solid- phase peptide synthesis using an automated microwave peptide synthesiser
- Custom DNA oligonucleotide synthesis
- Computer modelling for small molecule ligand design
- Fluorescent labelling of small molecule ligands and peptides Novel design of transition metal luminescent probes for imaging in cells
- Novel contrast agents for magnetic resonance imaging (MRI)
Physical measurement
- Customised spectroscopy strategies for new assay methodologies
- Design of fluorescence and label-free based biosensors
- Label-free imaging and sensing
- Micro fluidic strategies: including digital droplet single cell analysis, or single molecule quantification
- Optical manipulation of live cells
- Biological force measurements: mechanochemical cell and single molecule biology
- Raman and fluorescence confocal microspectroscopy
- Computer modelling: quantum mechanical ab initio calculations and classical molecular modelling
Super Resolution Microscopy
 Researchers at Leicester are developing cutting-edge super-resolution localisation microscopy. This technology breaks the fundamental diffraction limit of biological microscopes and permits imaging of biological reactions at molecular scale (10nm), in physiological conditions.
Researchers at Leicester are developing cutting-edge super-resolution localisation microscopy. This technology breaks the fundamental diffraction limit of biological microscopes and permits imaging of biological reactions at molecular scale (10nm), in physiological conditions.
It exploits switching of fluorescent molecules and to localize, and co-localize individual bio- molecules to within about 10nm. Switching of molecules can be either by stochastic (STORM) or deterministic (STED) blinking, with STORM and STED being the most common super- resolution modalities.
At Leicester, stochastic switching is achieved by binding of single molecules (protein or DNA) to their interacting partners immobilized on glass, in DNA nanoreactors, or inside living cells.
The microscope in development in the Henry Wellcome Building has the ability to collect four colours simultaneously (most single-molecule microscopes have two colours) for extended periods of time (hours as opposed to seconds and minutes) in the absence of artefacts due to drift (Lestascope drifts at 2 nm per hour, whereas commercial microscopes drift 2 μm per hour). This exciting technological development combined with the know-how of chemical labelling and surface chemistry from our researchers provides for the analysis of complex enzymatic mixtures.
NMR Data Analysis Software (CCPN)
 The Collaborative Computational Project for NMR (CCPN) is an international consortium of 28 partners which is working to provide the tools and knowledge to maximise the impact of the biological NMR studies. CCPN is chaired by Professor Geerten Vuister. The CCPN software facilitates data analysis and software integration, and the project actively promotes the exchange of knowledge, provides training and supports best practices in the NMR community.
The Collaborative Computational Project for NMR (CCPN) is an international consortium of 28 partners which is working to provide the tools and knowledge to maximise the impact of the biological NMR studies. CCPN is chaired by Professor Geerten Vuister. The CCPN software facilitates data analysis and software integration, and the project actively promotes the exchange of knowledge, provides training and supports best practices in the NMR community.
The latest release of the CCPN software package, Analysis-V3, presents a complete write of the entire code-base. The programs now provide a fast and smooth interface with drag-and-drop style operations and two-key shortcut interactions and easy data import and export, including .xlsx/.xls spreadsheet files that can contain any meta-data, such as chemical properties, including SMILES (Simplified molecular-input line-entry system). The software natively reads all common NMR data formats, including Bruker, Agilent, NmrPipe and ucsf formats and is fully NEF compatible.
The complexities in code and data structure are encapsulated under a layer of wrapper code, which enables users to write extensions to the software very quickly and simply. In addition, there is direct access to extensive scientific libraries, such PySci, Scikit-learn, which greatly facilitate data analysis using modern advanced technologies, such as machine-learning.
Protein Expression Laboratory (PROTEX)

The Protein Expression Laboratory (PROTEX) has been established to provide a high throughput service for cloning and small-scale protein expression trials. The laboratory is designed to serve the needs of structural biology, enzymology and molecular and cell biology including genome editing.