Leicester Microbial Sciences and Infectious Diseases Centre (LeMID)
Structural biology
Work in the College of Life Sciences on the structure of proteins (using techniques such as NMR spectroscopy and x-ray crystallography) are providing insight on a wide range of fundamental processes such as cell motility, drug metabolism, gene activation and tuberculosis virulence.
Defining protein structures are fundamental to understanding pathogenesis of microbes in addition to host response mechanisms. The study of structure is highly relevant to the study of microbiology and therefore the relationship with the Leicester Institute of Structural and Chemical biology is a valued collaboration.
- Streptococcus pneumoniae
- Tuberculosis
- Facilities
Streptococcus pneumoniae
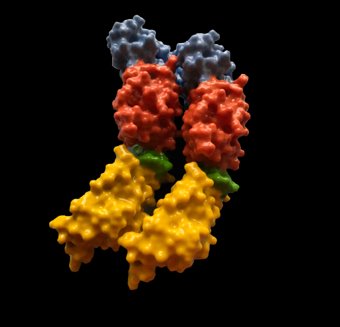
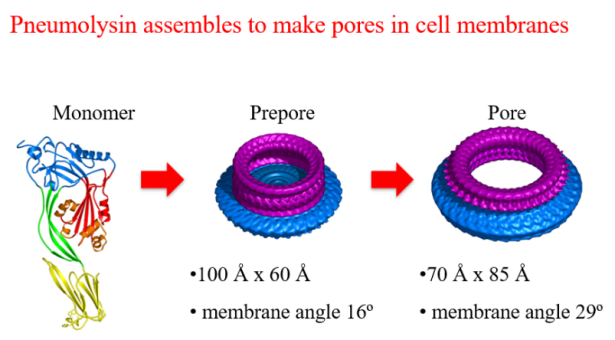
The sophisticated work of Dr P Andrew has defined the structure of pneumolysin, a S. pneumoniae toxin that is contrived of 4 domains. These domains assemble at the surface of the target mammalian cell and create a membrane attack complex.
This advancement in knowledge leads the way for investigation of novel vaccines and treatments at the university.
Pneumolysin is the toxin that disrupts cell membranes to allow the pneumococcus to attack mammalian cells. In 2015 a team led by Professor Peter Andrew and Professor Russell Wallis from our Department of Respiratory Sciences used X-ray crystallography to study pneumolysin at an eye-watering resolution of just 1.98 angstroms and from this they were able to achieve the long-sought goal of determining its molecular structure. Discovering how pneumolysin binds to membranes at a molecular level, means there is a possibility of designing a drug that can inactivate the pneumolysin.
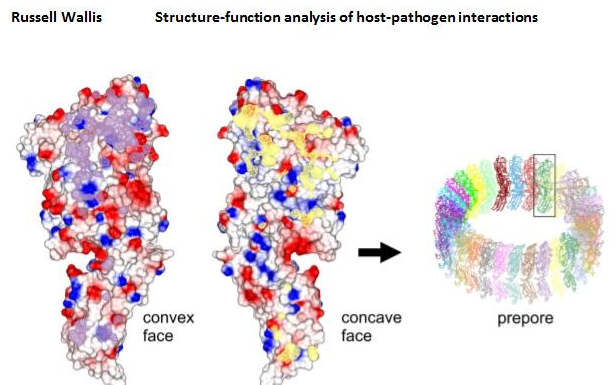
Furthermore, identifying structures are important in developing novel diagnosis techniques such as that of Dr J. T. Hodgkinson’s work on chemical probes.
The structure of pneumolysin, a pore-forming toxin from Streptococcus pneumoniae. Monomers pack side-by-side in the crystal in the same way as occurs on the surface of a mammalian cell prior to pore formation (the prepore). The interface between two monomers is shown in purple and yellow.
Structure and Tuberculosis
Bacterial signalling proteins
Dr Helen O’Hare’s team study PknG signalling in Mycobacterium tuberculosis, recent findings have elucidated sensor proteins within the pathways, contributing to the understanding of the bacteria’s metabolic pathway. They went on to identify the structure of the protein through X-ray crystallography at the Diamond Light Source, Harwell. Dr O’Hare stated:
“Our findings have broader significance for other Actinobacterial pathogens, such as non-tuberculous Mycobacteria, as well as Actinobacteria used to produce billions of dollars of amino acids and antibiotics every year”
- Read the research article ‘An aspartate-specific solute-binding protein regulates protein kinase G activity to control glutamate metabolism in Mycobacteria.'
Drug targets
Collaboratively, the LeMID network are working to validate anti-TB drug targets and for structure based drug discovery (Russell Wallis), to study protein phosphorylation at the level of the phosphoproteome and in regulation of bacterial cell growth (Galina Mukamolova), and to study the function of these signalling pathways in infection models (Leicester TB Research Group).
Quorum sensing
This natural phenomenon termed quorum sensing (QS) is achieved by the release of small signalling molecules which can diffuse between bacterial cells and can be detected by specific receptor proteins. Many phenotypes under QS regulation play an important role in bacterial virulence and directly impact pathogenicity of bacteria to eukaryotic hosts, as well as antibiotic tolerance. Dr Hodgkinson's research involves the design, synthesis and biological evaluation of natural and non-natural signalling molecules for structure activity relationship studies, mode of action studies and inhibitor design.