Virtual Genetics Education Centre
Antibiotic resistance for higher education
Antibiotics have successfully been used to treat bacterial infections for decades. Their use however, is becoming threatened as bacteria evolve mechanisms to become resistant to these drugs.
Antibiotics are molecules with activity against living organisms. When discussing antibiotics, we are often referring to antibacterials or drugs that kill bacteria. Antibiotics work by targeting and inhibiting key processes in a bacterium's biochemistry. For example, several classes of antibiotics (aminoglycosides and macrolides for example) work by inhibiting bacterial protein synthesis.
Following the discovery of penicillin we enjoyed a 'golden age' of medicine whereby nearly all bacterial diseases could be treated with relative ease. This is now hampered by the evolution of bacteria that can resist antibiotics, and the propagation of these bacteria through improper use of the drugs.
Mechanisms of resistance
There are 3 main mechanisms by which bacteria can become resistant to antibiotics. Better understanding these mechanisms is a key area of research.
(1) Changing the drug
Many bacteria code for enzymes that can modify antibiotics. The result is that often they can no longer bind to their target site. Beta-lactam antibiotics for example are degraded by a class of enzymes called beta-lactamases, which cleave the characteristic lactam ring of the drug rendering them inactive.
Genes encoding these elements can be acquired horizontally, but many bacteria encode these elements intrinsically. Mutations in these genes, or the promoter regions of these genes have been well characterised as a leading cause of clinical levels of resistance through increased gene expression.
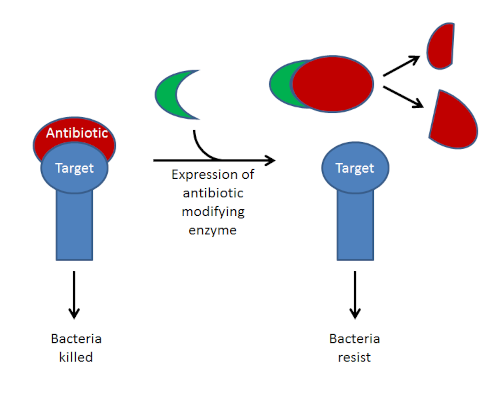
Above- this diagram depicts how expression of antibiotic-modifying enzymes can lead to drug resistance. In addition to enzymatic cleavage, some of these enzymes inactivate antibiotics by covalently linking them with other molecules, for example the aminoglycoside acetyl-transferases.
(2) Changing the target
In addition to drug modification, bacteria can become resistant by changing the shape of an antibiotic's target. These mutations often change the shape of the target, lowering the affinity to antibiotics, but not impairing their native function. Furthermore, bacteria can horizontally acquire decoy proteins with high affinities to antibiotic, meaning the drug will bind to these instead of their cellular target.
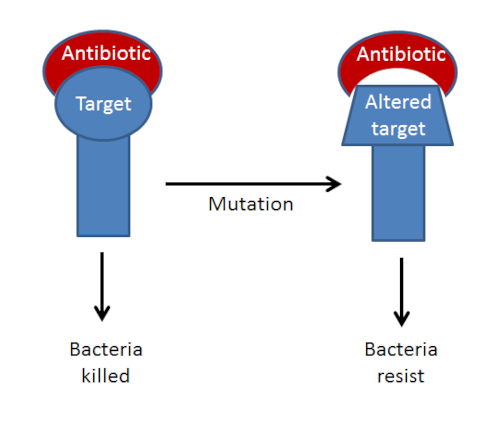
Above- this diagram shows mutation altering the structure of an antibiotic target. This is a common mechanism of resistance to quinolone antibiotics (such as ciprofloxacin). Quinlones target bacterial DNA gyrases and other type IV topoisomerases. Mutations in specific regions of the genes encoding these proteins (Quinolone Resistance Determining Region) have been shown to confer resistance.
(3) Active efflux
Finally, the mechanism most commonly associated with resistance to multiple classes of antibiotics (multi-drug resistance) are the action of efflux pumps. These multi-protein complexes can be very selective in their targets, or very broad. They continually pump antibiotics out of the cell, lowering intercellular concentrations of the drug, leading to resistance. Efflux pumps are often intrinsically present in susceptible strains implying a more general biological function. Mutations leading to overexpression of efflux pumps are a common feature in bacteria resistant to several classes of clinical antibiotics.
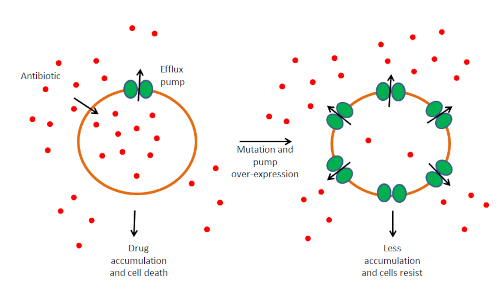
Above- The presence of efflux pumps in susceptible strains indicates that they have a function in the essential biology of the cell. Over expression of these pumps are what results in clinical levels of resistance.