People
Professor Daniel Panne
Professor in Structural Biology
School/Department: Molecular and Cell Biology, Department of
Email: daniel.panne@leicester.ac.uk
Research
Structural biology of signal transduction and epigenetic gene regulation
Our research has focused on understanding how signalling pathways control gene expression by epigenetic modulation of chromatin structure. One of the best-understood model systems for metazoan gene regulation is found in the innate immune system, which is crucial to limit pathogen infections: Several groups of pattern recognition receptors induce different signalling pathways leading to production of a variety of antiviral molecules including type I interferons and proinflammatory cytokines. Previous work from the laboratory has revealed mechanistic and structural insights into how the interplay between cellular signalling, TF activation and coassembly on transcriptional enhancers controls gene expression (Fig. 1). Higher-order transcription factor complexes such as the enhanceosome frequently recruit the co-activators CBP/p300. CBP/p300 are important to integrate the cellular signals by providing a scaffold function. CBP/p300 also acetylate chromatin and ultimately, in conjunction with remodellers and histone chaperones, makes chromatin permissive for gene transcription.
One important first step toward characterising such dynamic processes is to determine the molecular architecture of essential components. We are using a combination of biophysical techniques including X-ray crystallography, cryo-electron microscopy, crosslinking and native mass spectrometry, and more to address the following questions concerning information transfer in this system:
- What is the architecture of signalling complexes that direct innate immune responses and control gene expression?
- How do these signalling pathways lead to assembly of higher-order transcriptional regulatory complexes?
- How does assembly of such regulatory complexes ultimately lead to chromatin acetylation, a modification found on active chromatin?
- How does chromatin modification direct nucleosome remodelling and gene regulation?
- How do components that regulate higher order chromatin structure contribute to genome regulation?
Answers to some of these questions are expected to contribute to our understanding of chromatin regulation and dysregulation in disease. This is not only of fundamental importance for cellular signalling and gene regulation, but also opens up opportunities for pharmacological targeting.
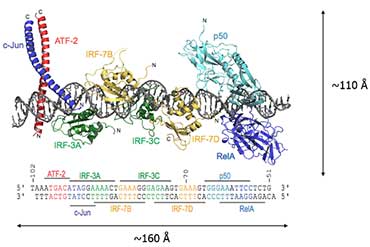
Diagram showing the assembly of an enhanceosome containing the transcription factors ATF-2/c-Jun, IRF-3/IRF-7, NF-kappaB and HMGI(Y) to control gene expression
Group members
Kelsey Hipwell, Mahima Patel, Connor Robertson, Amrita Patel, Gajanan Patil, Naveen Nagarakanti, Tajith Shaik
Publications
Key publications
-
Buenaventura T, Bagci H, Patrascan I, Graham JJ, Hipwell KD, Oldenkamp R, King JWD, Urtasun J, Young G, Mouzo D, Gomez-Cabrero D,Rowland BD, Panne D, Fisher AG, Merkenschlager M. (2024) Competition shapes the landscape of X-chromosome-linked genetic diversity. Nat Genet https://doi.org/10.1038/s41588-024-01840-5
- Muir K.W., Li Y., Weiss F., Panne D. (2020) The structure of the cohesin ATPase elucidates the mechanism of SMC-kleisin ring opening. Nat Struct Mol Biol 27: 233-239E.
- Ortega, S. Rengachari, Z. Ibrahim, N. Hoghoughi, J. Gaucher, A. S. Holehouse, S. Khochbin, D. Panne. (2018) 'Cellular signalling activates the p300 acetyltransferase through transcription factor dimerization.' Nature 562, pages 538-544
- Sauer P, et al. (2017) 'Insights into the molecular architecture and histone H3-H4 deposition mechanism of the Chromatin assembly factor 1'. eLife
- Kaczmarska Z et al. (2016) 'Structure of the p300 in complex with Acyl-CoA variants.' Nat. Chem. Biol, vol. 13. Pp. 21-29
- Aguilar–Gurrieri C.et al. (2016) 'Structural evidence for Nap1–dependent H2A–H2B deposition and nucleosome assembly.' EMBOJ, vol. 35. Pp. 1465-82