People
Professor John Schwabe
Professor of Structural Biology

School/Department: Molecular and Cell Biology, Department of
Email: john.schwabe@le.ac.uk
Profile
Professor John Schwabe’s research spans a number of interrelated areas concerned with understanding enzymes inside the cell nucleus that assemble into large complex molecular machines to regulate genes. His group has led the field in how these complexes are assembled, how they act together with other proteins to control physiology and how this mechanism is affected by disease.
The Institute of Structural and Chemical Biology houses the state-of-the-art £6 million Regional Cryo-Electron Microscopy facility, a revolutionary method to directly visualise the molecular machinery in our cells with atomic-level detail.
Research
- Structural Biology: NMR Spectroscopy and X-ray Crystallography
- Molecular mechanisms of the regulation of gene expression
- Nuclear receptors and their role in metabolism, development and disease
The current major interest in the group is understanding the structure and function of HDAC-containing corepressor complexes. This work is supported by a Wellcome Trust Senior Investigator Award. These complexes play an important role in the regulation of development, differentiation, cancer and homeostasis.
In addition, the group has a long-standing interest in the molecular mechanisms of transcriptional regulation by nuclear receptors. Nuclear receptors are important regulators of gene expression and are major drug targets. Our understanding of the mechanism of the ligand-regulated switch has been greatly enhanced by studies of the structure and dynamics of nuclear receptors.
In addition to these core research areas, the group is also involved in a number of collaborative projects with an emphasis on proteins and complexes implicated in various cancers.
Structural biology of transcriptional repression complexes
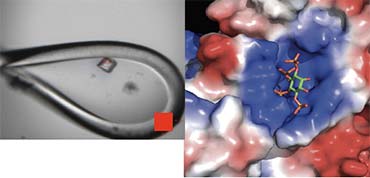 The major focus of my research group is to understand the molecular mechanisms that underlie the epigenetic reprogramming of the genome during cellular differentiation and development. Our particular interest is in Histone Deacetylase (HDAC) complexes whose primary role is to remove acetyl groups from lysine sidechains within the tails of histone proteins. This results in hypo-acetylated chromatin, which is the first step in gene inactivation.
The major focus of my research group is to understand the molecular mechanisms that underlie the epigenetic reprogramming of the genome during cellular differentiation and development. Our particular interest is in Histone Deacetylase (HDAC) complexes whose primary role is to remove acetyl groups from lysine sidechains within the tails of histone proteins. This results in hypo-acetylated chromatin, which is the first step in gene inactivation.
The Class 1 HDACs form the catalytic core of six families of HDAC-containing complexes that are targeted to chromatin. These contain the class I HDACs 1, 2 or 3 along with a variety of other proteins that target the complex to chromatin and/or possess other enzymatic activities. Importantly, the enzymatic activity of the HDACs requires assembly into these complexes.
These complexes are promising drug targets for the treatment of multiple disorders including cancer, Alzheimer’s Disease and HIV. To date at least six drugs which target HDACs are licensed for use in humans to treat certain types of cancer. Currently these drugs lack true isoform and complex specificity and therefore have adverse side-effects. By understanding the assembly and architecture of these complexes we hope to be able to develop drugs that are specific for single complexes.
We take an integrated structural biology approach to seek to understand this diverse family of HDAC complexes. We have recently made several major advances using X-ray crystallography techniques and we are now combining these studies with techniques such as negative-stain and cryo-electron microscopy, small-angle X-ray scattering and cross-linking mass spectrometry. These approaches can overcome the limitations of X-ray crystallography in studying large and somewhat flexible complexes. Using the information from structural biology we then utilise a wide variety of complementary techniques from biophysical to cell biology assays to fully understand their structure and function. We collaborate with groups from within the Institute and Department of Molecular and Cell Biology to develop chemical tools and probes (James Hodgkinson), to understand the function of these complexes in cells and animal models (Shaun Cowley / Andrew Fry).
See more of Professor John Schwabe's research highlightsPublications
Fox, J.L., Hughes, M.A., Powley, I.R., Juke-Jones, R., Ragan, T.J., Dinsdale, D., Fairall, L., Schwabe, J.W.R., Morone, N., Cain, K., MacFarlane, M. (2021) Cryo-EM structural analysis of FADD:Caspase-8 complexes defines the catalytic dimer architecture for coordinated control of cell fate. Nature Communications In Press.
Xiao, X., Kim, Y., Romartinez-Alonso, B., Sirvydis, K., Ory, D., Schwabe, J.W.R., Jung, M.E. and Tontonoz, P. (2021) Small molecule Aster inhibitors distinguish vesicular and non-vesicular sterol transport mechanisms. Proceedings of the National Academy of Sciences USA 118, e2024149118.
Turnbull, R., Fairall, L., Saleh, A., Kesall, E., Morris, K., Ragan, T., Savva, C., Chandru, A., Millard, C., Makarvova, O., Smith, C., Rosemann, A., Fry, A., Cowley, S., Schwabe, J.W.R. (2020) The MiDAC histone deacetylase complex is essential for embryonic development and has a unique multivalent structure. Nature Communications 11, 3252.
Millard, C.J., Fairall, L., Ragan, T.J., Savva, C.G. and Schwabe, J.W.R. (2020) The topology of chromatin-binding domains in the NuRD deacetylase complex. Nucleic Acids Research 48, 12972.
Song Y., Dagil l., Fairall L., Robertson N., Wu M., Ragan T.J., Savva G.C., Morone N., Kunze M.B.A., Jamieson A.G., Cole P.A., Hansen D.F., Schwabe J.W.R. (2020) Functional and structural coupling between LSD1 and HDAC1 in the CoREST complex. Cell Reports 30, 2699-2711.
Wang, Z.A., Millard, C.J., Lin, C-L., Gurnett, J., Wu, M., Lee, K., Fairall, L., *Schwabe, J.W.R., *Cole, P.A. (2020) Diverse Nucleosome Site-Selectivity Among Histone Deacetylase Complexes. eLife 9, e57663. *Co-corresponding authors
Smalley, J., Adams, G., Millard, C., Song, Y., *Schwabe, J.W.R., *Cowley, S., *Hodgkinson, J. (2020) PROTAC mediated degradation of Class-I histone deacetylase enzymes in corepressor complexes. Chemical Communications doi: 10.1039/d0cc01485k. *Co-corresponding authors
Williams, G.M., Paschalis, V., Ortega, J., Muskett, F.W., Hodgkinson, J.T., Li, G-M., Schwabe, J.W.R. and Lahue, R.S. (2020) HDAC3 deacetylates the DNA mismatch repair factor MutSb to stimulate triplet repeat expansions. Proceedings of the National Academy of Sciences USA 117, 23597.
Moran, C., Seger, C., Taylor, K., Oddy, O., Burling, K., Rajanayagam, O., Fairall, L., McGowan, A., Lyons, G., Halsall, D., Gurnell, M., Schwabe, J., Chatterjee, K., Strey, C. (2020) Hyperthyroxinemia and hypercortisolemia due to familial dysalbuminemia. Thyroidhttps://doi.org/10.1089/thy.2020.0315.
Freeman S.L., Portolano N., Parkin G., Girija U.V., Kwon H., Basran J., Fielding A.J., Fairall L., Svistunenko D.A., Moody P.C.E., Schwabe J.W.R., Kyriacou C.P. and Raven E.L. (2019) Heme Binding to human CLOCK affects interactions with the E-box. Proceedings of the National Academy of Sciences USA 116, 19911.
See more of Professor John Schwabe's publications via ResearchGate