Virtual Genetics Education Centre
Recombinant DNA and genetic techniques for schools and colleges
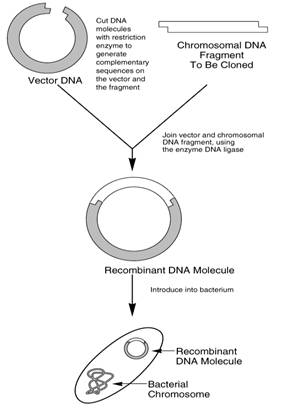
Recombinant DNA (or rDNA) is made by combining DNA from two or more sources. In practice, the process often involves combining the DNA of different organisms. The process depends on the ability to cut and re-join DNA molecules at points which are identified by specific sequences of nucleotide bases called restriction sites. DNA fragments are cut out of their normal position in the chromosome using restriction enzymes (also called restriction endonucleases) and then inserted into other chromosomes or DNA molecules using enzymes called ligases.
Gene cloning
This describes the process of copying fragments of DNA which can then be used for many different purposes, such as creating GM crops, or finding a cure for disease. There are two types of gene cloning: in vivo, which involves the use of restriction enzymes and ligases using vectors and cloning the fragments into host cells (as can be seen in the image above). The other type is in vitro which is using the polymerase chain reaction (PCR) method to create copies of fragments of DNA.
For in vivo cloning a fragment of DNA, containing a single gene or a number of genes, is inserted into a vector that can be amplified within another host cell. A vector is a section of DNA that can incorporate another DNA fragment without losing the capacity for self-replication, and a vector containing an additional DNA fragment is known as a hybrid vector. If the fragment of DNA includes one or more genes the process is referred to as gene cloning.
There are 4 different type of vectors:
- Plasmid vectors
- Lamda (λ) phage vectors
- Cosmids
- Expression vectors
The host cell copies the cloned DNA using its own replication mechanisms. A variety of cell types are used as hosts, including bacteria, yeast cells and mammalian cells.
Polymerase Chain Reaction (PCR)
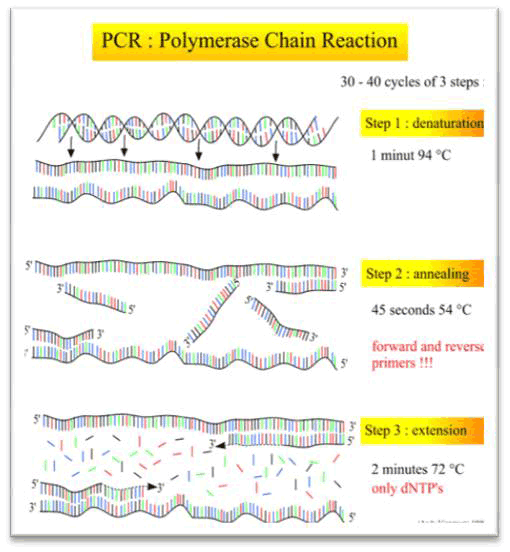
This is an in vitro method for making many copies of a specific section of DNA, without the need for vectors or host cells. The DNA to be copied – the template DNA – is mixed with forward and reverse primers complementary to the end of the template DNA, nucleotides, and a version of DNA polymerase known as Taq polymerase. (This enzyme is stable under high temperatures, and is obtained from the thermophilic bacterium Thermus aquaticus.) The process involves the repetition of three steps:
- denaturation, which separates the two nucleotide strands of the DNA molecule
- primer annealing, in which the primers bind to the single-stranded DNA
- extension, in which nucleotides are added to the primers – in the 5' to 3' direction – to form a double-stranded copy of the target DNA.
Each cycle takes a few minutes, and repeated cycles can produce large amounts of a specific DNA sequence in a matter of hours rather than days. However, this cloning method does require knowledge of some details about the nucleotide sequence to be copied, and the technique is very sensitive to small amounts of contamination.
Gene libraries
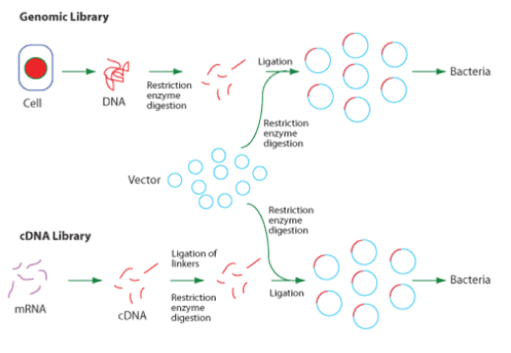
A gene library is a large collection of cloned DNA sequences from a single genome. A genomic library, (as can be seen above) in theory, would contain at least one copy of every sequence in an organism’s genome. These are used to investigate the structure of a given chromosome, or to clone specific genes. These types of libraries may be prepared from a subset of the entire genome (for example, a single chromosome). The first step in creating a genomic library is to break up, or ‘fractionate’, the genome using physical methods or restriction enzymes. The fragments are then linked to appropriate vectors and cloned in a suitable host cell population.
A cDNA library (complementary DNA) contains DNA present in a given cell population which is prepared from the mRNA (messenger RNA) using the enzyme reverse transcriptase. The resulting cDNA represents the genes expressed in the cell population as a subset of the entire genome, and can be cloned using a vector and suitable host cell (as seen in the diagram above). The cDNA will not include introns or regulatory sequences as these are removed from the RNA during processing, and this makes a cDNA library easier to maintain. A cDNA library can also be prepared using reverse transcriptase PCR (RT-PCR).
The identification of gene products in a gene library
Restriction enzymes (to cut the DNA) and gel electrophoresis (to separate the resulting fragments) can be used to produce a physical map of DNA segments in a process known as restriction mapping.
There are also a number of techniques that can be used to identify specific genes or gene products within a gene library and these are: Southern blotting, Northern blotting and Western blotting. However, the most powerful experimental technique for investigating genetics at the molecular level is DNA sequencing, which allows the nucleotide sequences of genes – even whole chromosomes – to be determined. Automated sequencing technologies are now allowing us to sequence the entire genomes of organisms from bacteria to human beings.
Molecular genetics and biotechnology
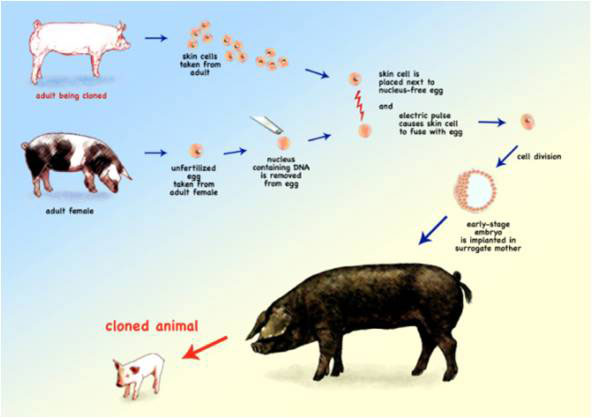
The new techniques of molecular genetics, combined with developments in associated biotechnologies, have led to advances in a number of different fields. We can now analyse the genomes of species that make an important contribution to agriculture, fuel production or drug development. We can move specific genes from one organism to another to create transgenic plants and animals, and use animal cloning techniques to produce animals that are genetically identical, such as Dolly the sheep, and more recently, cloned pets such as cats and dogs.
The process of cloning is straightforward, but the results are not always predictable. It took many hundreds of attempts to get it to work and produce one live sheep, and cloning in itself raises many questions not only about benefits and risks but also many ethical questions.
The technique of genetic fingerprinting, which enables the identification of individuals and the relationships between individuals has found many applications in science today. There is also ongoing research into gene therapy which examines the possibility of introducing cloned genes to compensate for defective, mutant genes. And other areas, for example, human cloning and stem cell research open up many ethical issues that must be addressed alongside the scientific developments.