People
Dr Yolanda Markaki
Lecturer
School/Department: Molecular Cell Biology, Department of; LISCB
Research
Developmental Epigenetics and Nuclear Architecture

Research highlights
Genome organization and function; epigenetic regulation of pluripotency and differentiation; heterochromatin formation; biomolecular assembly in gene regulation; X-chromosome inactivation; long non-coding RNAs; quantitative super-resolution imaging of nuclear landscapes
Research summary
Nuclear architecture plays a key role in the regulation of nuclear function and gene expression. Chromatin is organized into intricate specialized neighborhoods within the cell nucleus. These chromatin compartments arise from the local concentration of epigenetic factors and other effector molecules, including non-coding RNAs and architectural proteins, that modulate chromatin structure and gene expression. The three-dimensional organization and composition of our epigenomes is a dynamic process that changes during development or in disease states.
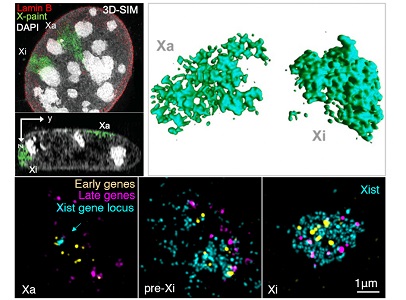 Our research group is interested in deciphering fundamental principles that govern genome architecture and to explore how changes in nuclear compartmentalization and the distribution of epigenetic factors alter gene expression and cellular fate. To understand epigenetic mechanisms, we primarily use X chromosome inactivation (XCI) as a model system. XCI is a fundamental gene regulatory process that takes place in early embryonic development to compensate the imbalance of X-linked gene dosage between XX females and XY males. Once established, the inactive X will be clonally propagated in daughter cells as heritable epigenetic memory. Both active and inactive X chromosome territories carry the same genetic code, yet their structural, molecular and gene expression profiles are profoundly different. Initiation of XCI is essential for the survival and development of female embryos while its maintenance in adult tissues is required to regulate cell identity and physiology. Its dysregulation leads to severe diseases, including immune diseases and cancer by partial or complete X chromosome reactivation.
Our research group is interested in deciphering fundamental principles that govern genome architecture and to explore how changes in nuclear compartmentalization and the distribution of epigenetic factors alter gene expression and cellular fate. To understand epigenetic mechanisms, we primarily use X chromosome inactivation (XCI) as a model system. XCI is a fundamental gene regulatory process that takes place in early embryonic development to compensate the imbalance of X-linked gene dosage between XX females and XY males. Once established, the inactive X will be clonally propagated in daughter cells as heritable epigenetic memory. Both active and inactive X chromosome territories carry the same genetic code, yet their structural, molecular and gene expression profiles are profoundly different. Initiation of XCI is essential for the survival and development of female embryos while its maintenance in adult tissues is required to regulate cell identity and physiology. Its dysregulation leads to severe diseases, including immune diseases and cancer by partial or complete X chromosome reactivation.
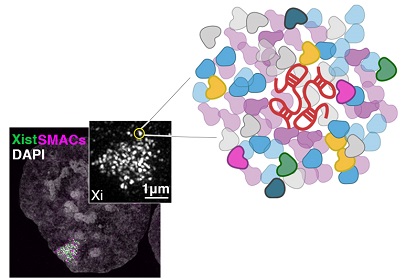 The long non-coding (lnc) RNA Xist triggers XCI by recruiting to the inactive X an array of epigenetic factors and other effector proteins that induce higher-order chromatin changes and transcriptional repression. Our work has uncovered that Xist forms nanosized supramolecular complexes (SMACs) that regulate the initiation of silencing on the inactive X. SMACs arise from only few RNA molecules, form novel protein-protein interactions that are essential to their function and regulate broad genomic regions. A major unanswered question is how these RNA-seeded supercomplexes change and mature to initiate and maintain transcriptional shutdown during development and then how these are disrupted in pathologies. To address these questions we integrate genetic engineering, cell and molecular biology approaches with cutting-edge quantitative super-resolution microscopy. We use several stem cell and disease models in collaboration with the University Hospitals Leicester and the Leicester Cancer Research Center to understand how changes in genome organization and the composition of epigenetic factors contribute to different expression patterns in development or disease. We collaborate with other groups within the Institute and the Department of Molecular and Cell Biology to obtain a structure-function relationship of epigenetic complexes and protein-protein interactions critical to their function.
The long non-coding (lnc) RNA Xist triggers XCI by recruiting to the inactive X an array of epigenetic factors and other effector proteins that induce higher-order chromatin changes and transcriptional repression. Our work has uncovered that Xist forms nanosized supramolecular complexes (SMACs) that regulate the initiation of silencing on the inactive X. SMACs arise from only few RNA molecules, form novel protein-protein interactions that are essential to their function and regulate broad genomic regions. A major unanswered question is how these RNA-seeded supercomplexes change and mature to initiate and maintain transcriptional shutdown during development and then how these are disrupted in pathologies. To address these questions we integrate genetic engineering, cell and molecular biology approaches with cutting-edge quantitative super-resolution microscopy. We use several stem cell and disease models in collaboration with the University Hospitals Leicester and the Leicester Cancer Research Center to understand how changes in genome organization and the composition of epigenetic factors contribute to different expression patterns in development or disease. We collaborate with other groups within the Institute and the Department of Molecular and Cell Biology to obtain a structure-function relationship of epigenetic complexes and protein-protein interactions critical to their function.
Publications
Dror I, Chitiashvili T, Tan SYX, Cano CT, Sahakyan A, MarkakiY, Chronis C, Collier AJ, Deng W, Liang G, Sun Y, Afasizheva A, Miller J, Wen Xiao, Douglas L. Black, Ding F, Plath K. XIST directly regulates X-linked and autosomal genes in naive human pluripotent cells. Cell. 187, 1, doi:/10.1016/j.cell.2023.11.033
Markaki Y*, Chong JG, Wang Y, Jacobson EC, Luong C, Tan SYX, Jachowicz JW, Strehle M, Maestrini D, Dror I, Mistry BA, Schöneberg J, Banerjee A, Guttman M, Chou T*, Plath K*. Xist nucleates local protein gradients to propagate silencing across the X chromosome. Cell. 2021 doi: 10.1016/j.cell.2021.10.022. *co-corresponding authors
Pandya-Jones A, Markaki Y, Serizay J, Chitiashvili T, Mancia Leon WR, Damianov A, Chronis C, Papp B, Chen CK, McKee R, Wang XJ, Chau A, Sabri S, Leonhardt H, Zheng S, Guttman M, Black DL, Plath K. A protein assembly mediates Xist localization and gene silencing. Nature. 2020;587(7832):145-51. doi: 10.1038/s41586-020-2703-0
Kraus F, Miron E, Demmerle J, Chitiashvili T, Budco A, Alle Q, Matsuda A, Leonhardt H, Schermelleh L, Markaki Y. Quantitative 3D structured illumination microscopy of nuclear structures. Nat Protoc. 2017;12(5):1011-28. doi: 10.1038/nprot.2017.020
Anton T, Bultmann S, Leonhardt H*, Markaki Y*. Visualization of specific DNA sequences in living mouse embryonic stem cells with a programmable fluorescent CRISPR/Cas system. Nucleus. 2014;5(2):163-72. doi: 10.4161/nucl.28488. *co-corresponding author