People
Dr Ralf Schmid
Associate Professor in Bioinformatics
School/Department: Molecular Cell Biology, Department of
Email: rs206@leicester.ac.uk
Profile
My research group has a broad interest in the application and development of computational methods covering different aspects of molecular modelling and bioinformatics. Techniques used in our lab are protein structure prediction by homology modelling and ab-initio modelling, ligand docking and virtual screening, molecular dynamics simulations and free energy calculations, evolutionary analysis of protein families, and a range of sequence based bioinformatics methods.
Research
Molecular modelling and bioinformatics
My research group has a broad interest in the application and development of computational methods covering different aspects of molecular modelling and bioinformatics. Techniques typically used in our lab are protein structure prediction by homology modelling and ab-initio modelling, ligand docking and virtual screening, molecular dynamics simulations and free energy calculations, evolutionary analysis of protein families, and a range of bioinformatics methods.
A main focus in our recent work is on understanding the molecular basis of P2X receptor activation and antagonism (in collaboration with Prof Evans’ group, University of Leicester). P2X receptors are ligand-gated cation channels activated by binding of extracellular ATP. The human genome encodes seven paralogs of the P2X receptor protein family (P2X1R-P2X7R) which form homo- and hetero-trimers. P2X receptors are involved in a variety of physiological and pathological processes, and specifically P2X7R is established as a drug target. For P2X7R we are interested in the features that determine paralog specificity of steric and allosteric antagonists, and we have developed a robust protocol for predicting binding poses of P2X7R antagonists. Another project is focused on determining conformational changes occurring in the transition between different states of P2X receptors, and we calculate structural models for the P2XR1 intracellular domain using ab-initio modelling informed by crosslinking derived distance constraints.
Other areas of work are in studying the evolution of function and binding sites in protein families by phylogenetic analysis, using ligand docking and molecular dynamics simulations to investigate kinase activation and inhibition, developing methods for the functional annotation of protein sequences, and studying alternative splicing and its effect on protein structure based on RNAseq data.
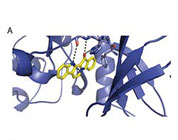
Molecular model for fascaplysin bound to CDK4.
Molecular model for surfaces of P2X1 receptor.

Phylogenetic alignment of phospho protein family.
Publications
Discovery and structure relationships of salicylanilide derivatives as potent, non-acidic P2X1 receptor antagonists
M Tian, A Abdelrahman, Y Baqi, E Fuentes, D Azazna, C Spanier, S Densborn, S Hinz, R Schmid, CE Müller (2020)
Journal of medicinal chemistry 63 (11), 6164-6178
Mapping the site of action of human P2X7 receptor antagonists AZ11645373, Brilliant Blue G, KN-62, Calmidazolium, and ZINC58368839 to the intersubunit allosteric pocket
AB Dayel, RJ Evans, R Schmid (2019)
Molecular pharmacology 96 (3), 355-363
ATP-gated P2X receptor channels: molecular insights into functional roles
R Schmid, RJ Evans (2019)
Annual review of physiology 81, 43-62
A mechanism for CO regulation of ion channels
SM Kapetanaki, MJ Burton, J Basran, C Uragami, PCE Moody, JS Mitcheson, R Schmid, NW Davies, P Dorlet, MH Vos, NM Storey, E Raven (2018)
Nature communications 9 (1), 1-10
Unique residues in the ATP gated human P2X7 receptor define a novel allosteric binding pocket for the selective antagonist AZ10606120
RC Allsopp, S Dayl, R Schmid, RJ Evans (2017)
Scientific reports 7 (1), 1-12 35
A heme-binding domain controls regulation of ATP-dependent potassium channels
MJ Burton, SM Kapetanaki, T Chernova, AG Jamieson, P Dorlet, J Santolini, PCE Moody, JS Mitcheson, NW Davies, R Schmid, EL Raven, NM Storey (2016)
Proceedings of the National Academy of Sciences 113 (14), 3785-3790
The splicing landscape is globally reprogrammed during male meiosis
R Schmid, SN Grellscheid, I Ehrmann, C Dalgliesh, M Danilenko, MP Paronetto, S Pedrotti, D Grellscheid, RJ Dixon, C Sette, IC Eperon, DJ Elliott (2013)
Nucleic acids research 41 (22), 10170-10184
Fascaplysin as a specific inhibitor for CDK4: Insights from molecular modelling
MI Shafiq, T Steinbrecher, R Schmid (2012)
Public Library of Science 7 (8), e42612
Agonist binding evokes extensive conformational changes in the extracellular domain of the ATP-gated human P2X1 receptor ion channel
JA Roberts, RC Allsopp, S El Ajouz, C Vial, R Schmid, MT Young, RJ Evans (2012)
Proceedings of the National Academy of Sciences 109 (12), 4663-4667
Cytosolic metal handling in plants: determinants for zinc specificity in metal transporters and metallothioneins
CA Blindauer, R Schmid (2010)
Metallomics 2 (8), 510-529
annot8r: GO, EC and KEGG annotation of EST datasets
R Schmid, ML Blaxter (2008)
BMC bioinformatics 9 (1), 1-6
A transcriptomic analysis of the phylum Nematoda
J Parkinson, M Mitreva, C Whitton, M Thomson, J Daub, J Martin, R Schmid, N Hall, B Barrell, RH Waterston, JP McCarter, ML Blaxter (2004)
Nature genetics 36 (12), 1259-1267
PartiGene—constructing partial genomes
J Parkinson, A Anthony, J Wasmuth, R Schmid, A Hedley, M Blaxter (2004)
Bioinformatics 20 (9), 1398-1404
Supervision
Projects are available on molecular modelling of P2X receptors (see above). Beyond this the lab has a broad interest in evolution of protein families and more generally in how proteins work, and we study these questions by "Computational Biochemistry".
Teaching
Undergraduate teaching
- BS2040 Bioinformatics
- BS2093 Protein Control in Cellular Regulation
- BS3070 Structural Biology.
Postgraduate teaching (MSc in Bioinformatics)
- BS7102 Proteins: Structure and Bioinformatics
- BS7105 Bioinformatics Programming and Advanced Topics in Bioinformatics
- BS7120 Steered Research Project
Press and media
Molecular Modelling in Health and Disease
Qualifications
- Diplom Chemiker (sehr gut) Albert-Ludwigs-Universität Freiburg
- Dr. rer. nat. (summa cum laude) Albert-Ludwigs-Universität Freiburg
- FHEA