People
Professor Geerten Vuister
Professor in Structural Biology
School/Department: Molecular and Cell Biology, Department of
Email: gv29@leicester.ac.uk
Research
Structural biology of Ca2+-transport regulation complexes
The research in my group is focused in two main areas: Firstly, on protein domains involved in the regulation of ion transport and the regulation and assembly of active biological complexes. Secondly, on the development of methods to improve NMR-derived structures, develop automated tools for structure quality assessment and data evaluations, and design new NMR sequences for the study of dynamics and interactions.
A particular area of interest is understanding the regulatory mechanisms that govern Ca2+ fluxes across membranes, which are crucial in many cellular processes. The systems we are currently studying involves the Na+/Ca2+ exchanger (NCX), a highly ubiquitous ion transporter that constitutes the dominant Ca2+ efflux mechanism in heart and sensory neurons, and the family of Transient Receptor Potential (TRP) channels, in particular the vaniloid TRPV5/6 subtypes. We use high-resolution Nuclear Magnetic Resonance spectroscopy, together with other structural methods like SAXS and homology modelling, to understand the atomic detail underpinning the regulation of these channels. We then combine this information with data from biophysical and ultimately cell based assays to gain a comprehensive knowledge regarding the structure and function of these channels. These insights will hopefully aid the successful development of new drugs targeting the numerous devastating diseases associated with aberrant Ca2+ signalling, such as multiple sclerosis, heart disease, stroke, osteoporosis and Alzheimer’s disease.
For many years, my research group has been involved in the development of computational tools for both the analysis and validation of structural NMR data. The aim of these tools is to ensure that NMR structures adequately reflect the experimental data and are reliable in terms of overall and local quality. Currently, our efforts are focused within the Collaborative Computational Project for NMR (CCPN, MRC-funded). The aim of CCPN is to develop a graphics-based, adaptable and easily extendable software platform for users which covers all aspects of NMR data analysis from spectrum visualisation to structure calculation and validation. Through its active outreach programme, CCPN also promotes the exchange of knowledge, provides training, and supports best practices in the NMR community.
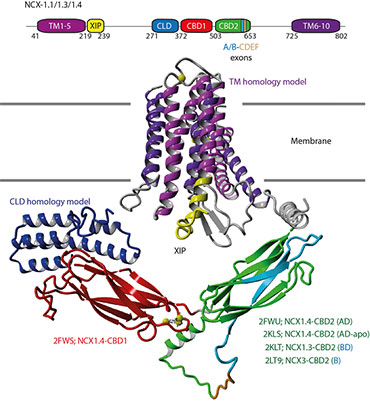
Molecular structure of the NCX protein showing its domain structure and membrane-binding domain.
Group members
Ed Brooksbank, Navaneet Chaturvedi, Vicky Higman, Luca Mureddu, Eliza Ploskon
Publications
Key publications
- Mureddu LG, Ragan TJ, Brooksbank EJ, Vuister GW. (2020) CcpNmr AnalysisScreen, a new software programme with dedicated automated analysis tools for fragment-based drug discovery by NMR. J Biomol NMR. doi: 10.1007/s10858-020-00321-1.
- Mureddu L and Vuister GW (2019) 'Simple high-resolution NMR spectroscopy as a tool in molecular biology.' FEBS J.;286(11):2035-2042.
- Bokhovchuk FM, Bate N, Kovalevskaya NV, Goult BT, Spronk CAEM, Vuister GW. (2018) The Structural Basis of Calcium- Dependent Inactivation of the Transient Receptor Potential Vanilloid 5 Channel. Biochemistry. 57(18):2623-2635.
- Gutmanas A, et al. (2015) 'NMR Exchange Format: a unified and open standard for representation of NMR restraint data.' Nat Struct Mol Biol, vol. 22. Pp. 433–434.
- Skinner SP, et al. (2016) CcpNmr AnalysisAssign: a flexible platform for integrated NMR analysis. J. Biomol. NMR 66: 111–124