People
Prof. Cyril Dominguez
Professor in Structural Biology
School/Department: Molecular Cell Biology, Department of
Telephone: +44 (0)116 229 7073
Email: cd180@leicester.ac.uk
Profile
I am a structural biologist interested mainly in protein-RNA interactions involved in the regulation of alternative splicing. I obtained my BSc and MSc in Biochemistry from the University of Aix-Marseille (France). In 2004, I obtained my PhD from the University of Utrecht (The Netherlands) under the supervision of Rob Kaptein Rolf Boelens and Alexandre Bonvin. During that time I developed a novel protein-protein docking software HADDOCK. After my PhD in 2004 I joined the laboratory of Fred Allain at the ETH Zurich (Switzerland) where I studied the interaction between the splicing factor hnRNP F and G-tract RNAs. In 2010 I obtained an MRC Career Development Award Fellowship to initiate my own research programme at the University of Leicester. My research focus on i) the interaction between the splicing factor Sam68 and its target RNA and ii) the role of RNA G-quadruplexes in the regulation of alternative splicing. In 2015 I became a lecturer, then an Associate Professor (2018) and a Professor (2023) at the University of Leicester.
Research
Alternative splicing of pre-mRNA is a centrally important and extraordinary cellular process that allows the generation of proteomic diversity from only 20000 human genes. The process of alternative splicing is very complex and still poorly understood. It is regulated by many proteins called splicing factors that may either enhance or prevent the recognition of a particular splice site in a time cell-type and cell-cycle dependent manner. Cell signalling also plays a major role in the regulation of alternative splicing through post-translational modifications of splicing factors demonstrating a tight link between cell signalling and alternative splicing regulation.
Our research interest is to understand at the molecular level the role of splicing factors on alternative splicing. Additionally we are interested in the role of pre-mRNA secondary and tertiary structures on alternative splicing regulation.
To this aim we are studying RNA protein-protein and protein-RNA complexes using structural biology techniques (NMR and X-ray) biophysical methods (ITC Fluorescence polarization) and biochemical methods (RNA footprinting EMSA).
Structure and function of RNA processing complexes
Alternative splicing of pre-mRNA is a centrally important and extraordinary cellular process that allows the generation of proteomic diversity (estimated at up to 150,000 human proteins) from only 20,000 human genes. More than 90% of human genes use multiple patterns of splicing, sometimes with antagonistic functions, to be expressed from a single gene. The process of alternative splicing is very complex and still poorly understood. The selection of particular splice sites among numerous potential sites is influenced by many proteins, called splicing factors, that bind to the pre-mRNA. These proteins may either enhance or prevent the recognition of a particular splice site in a time, cell-type and cell-cycle dependent manner. It has also been demonstrated that cell signalling plays a major role in the regulation of alternative splicing through post-translational modifications of splicing factors demonstrating a tight link between cell signalling and alternative splicing regulation.
Accordingly, pre-mRNA mutations as well as splicing factor mutations or copy number variations are associated with several genetic diseases including cancer.
Our main research interest is therefore to understand at the molecular level the specific action of splicing factors on alternative splicing decision, the interplay between signalling pathways and post-transcriptional gene regulation. Additionally, we are interested in the role of pre-mRNA secondary and tertiary structures on alternative splicing regulation.
To this aim, we are studying RNA, protein-protein and protein-RNA complexes using structural biology techniques (NMR and X-ray), biophysical methods (ITC, Flourescence polarization) and biochemical methods (RNA footprinting, EMSA,).
Our structural work is complemented by functional assays thank to collaboration with the group of Professor Ian Eperon within the LISCB.
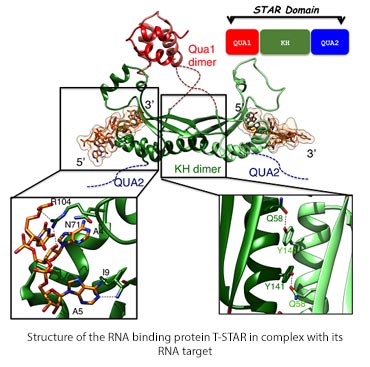
The diagram shows the structure of the dimeric RNA binding protein T-star in backbone only, highlighting the dimer interface with aromatic and glutamine sidechains and RNA binding site. Also a schematic showing the domain structure.
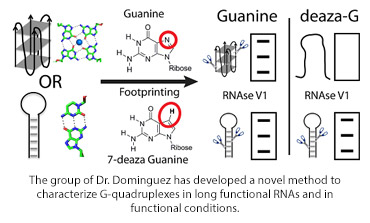
The diagram shows a footprinting method to distinguish between RNA quadruplexes and hairpin loops (shown schematically) comparing the effects of guanine and 7-deaza guanine (chemical structures shown) using RNAseIV, with the expected electrophoresis gels shown.
Publications
A potential histone-chaperone activity for the MIER1 histone deacetylase complex. Nucleic Acids Res., 51, 6006-19
Castelli, L.M, Lin, Y.H., Sanchez-Martinez, A., Gül, A., Imran, K.B., Higginbottom, A., Upadhyay, S.K., Márkus, N.M., Rua Martins, R., Cooper-Knock, J., Montmasson, C., Cohen, R., Walton, A., Bauer, C.S., De Vos, K.J., Mead, R.M., Azzouz, M., Dominguez, C., Ferraiuolo, L., Shaw, P.J., Whitworth, A.J., Hautbergue, G.M. (2023) A cell-penetrant peptide blocking C9ORF72-repeat RNA nuclear export suppresses neurodegeneration. Sci. Transl. Med., 15, eabo3823
Malki, M., Liepina I., Kogelnik, N., Watmuff, H., Robinson, S. Lightfoot, A., Gonchar, O., Bottrill, A., Fry, A.M., Dominguez, C. (2022) Cdk1-mediated threonine phosphorylation of Sam68 modulates its RNA binding, alternative splicing activity, and cellular functions. Nucleic Acids Res., 50, 13045-62.
Bueno-Alejo, et al. (2022) Surface passivation with a perfluoroalkane brush improves the precision of single molecule measurements. ACS Applied Material and Interfaces. 14, 49604-16
Bhogadia, M., Stone, B., Guerra, R. D. V., Muskett, F. W., Ghosh, S., Taladriz-Sender, A., .Burley, G.A., Eperon. I.C., Hudson, A.J., Dominguez, C. (2022). Biophysical characterisation of the Bcl-x pre-mRNA and binding specificity of the ellipticine derivative GQC-05: Implication for alternative splicing regulation. Front. Mol. Biosci., 9:943105
A. Ameerul , H. Almasmoum, L. Pavanello, C. Dominguez, G.S. Winkler (2022) Structural model of the human BTG2-PABPC1 complex by combining mutagenesis, NMR chemical shift perturbation data and molecular docking. J. Mol. Biol., 434, 167662
D. Munnur, J. Somers, G. Skalka, R. Weston, R. Jukes-Jones, M. Bhogadia, C. Dominguez, K. Cain, I. Ahel, M. Malewicz. (2019) NR4A Nuclear Receptors Target Poly-ADP-Ribosylated DNA-PKcs Protein to Promote DNA Repair. Cell Rep. 26:2028-2036
Weldon C,. Dacanay J.G., Gokhale V., Boddupally P.V.L., Behm-Ansmant I., Burley G.A., Branlant C., Hurley L.H., Dominguez C., Eperon I.C. (2018). Specific G-quadruplex ligands modulate the alternative splicing of Bcl-x. Nucleic Acids Res., 46, 886-96
Weldon C., Behn-Ansmant I., Burley G.A., Hurley L.H., Branlant C., Eperon I.C., and Dominguez C. (2016). Identification of G-quadruplexes using 7-deaza-guanine substituted RNA. Nat. Chem. Biol., 13, 18-20
M. Danilenko, C. Dalgliesh, V. Pagliarnini, C. Naro, I. Ehrmann, M. Feracci, M.K. Chadegani, A. Tyson-Capper, G.J. Clowry, P. Fort, C. Dominguez, C. Sette and D.J. Elliott (2017). Binding site density enables paralog-specific activity of SLM2 and Sam68 proteins in Neurexin2 AS4 splicing control. Nucleic Acids Res., 45(7), 4120-30.
Feracci M., Foot J.N., Grellscheid S.N., Danilenko M., Stehle R., Gonchar O., Kang H.S., Meyer N.H., Liu Y., Lahat A., Sattler M., Eperon I.C., Elliott D.J., and Dominguez C. (2016). Structural basis of RNA recognition and dimerization by the STAR proteins T-STAR and Sam68. Nat. Commun., 7, 10355
Supervision
Gene expression structural biology NMR
Teaching
- BS3070: Structural Biology. Co-convenor and Lecturer on NMR
- BS3010: Gene expression. Lecturer on protein-RNA interactions
- MB2050: Medical Biochemistry. Lecturer on Structure-based Drug design
- MSc Bioinformatics: Lecturer on Protein structure and NMR