Professor IC Eperon
Professor of Biochemistry
School/Department: Molecular Cell Biology, Department of
Telephone: +44 (0)116 229 7012
Email: eci@leicester.ac.uk
Profile
I studied Biochemistry at the University of Bristol from 1974 to 1977 and then worked for my PhD with Dr Fred Sanger at the MRC Laboratory of Molecular Biology Cambridge. This was followed by a year of post-doctoral work in the same laboratory. Following this I went to Yale for 2 years (1982-3) on an SERC-NATO fellowship to work with Professor Joan Steitz on RNA splicing. I began at Leicester as a lecturer in 1984 and was appointed as a Reader and then Professor of Biochemistry.
Research
Research group - RNA Splicing: from single-molecules to therapy
Gene expression in higher eukaryotes is dominated by two processes. Quantitative control of expression at any time is achieved by a range of processes, the dominant one of which is transcription. Qualitative control, the identity of what is expressed, is achieved by RNA splicing. Most genes produce multiple transcripts or isoforms as a result of splicing the initial transcript in different ways; some of these choices are tightly regulated and the encoded protein isoforms play essential roles in cell growth, development, circadian rhythms, memory etc.
The dramatic diversity of splicing patterns is achieved by weakening the standard signals for splicing and relying on the recognition of a very large number of short sequence motifs that are recognised by numerous activator or repressor proteins. This means that, for many genes, splicing involves an overwhelming number of choices and that the process of selection is very complex. Our work focuses on the mechanisms of selection and also on our ability to usurp these for therapeutic benefits.
We have made important contributions to identifying the roles of transcription, RNA folding and splice site sequences and the effects of regulatory proteins on the binding of core splicing components. Many of these were achieved using the one functional system available in vitro, a crude nuclear extract. However, we realised that the outcome of each event is the result of binding by numerous proteins, often in competition, and that the RNA-protein complexes being studied in extracts were likely to be very heterogeneous assemblages. To overcome this, we pioneered (in collaboration with Clive Bagshaw, Dmitry Cherny and Andrew Hudson) the use of single molecule methods in crude extracts, and we have used these to look at the numbers of molecules of particular proteins bound to each molecule of RNA and establish how this affects activation or repression.
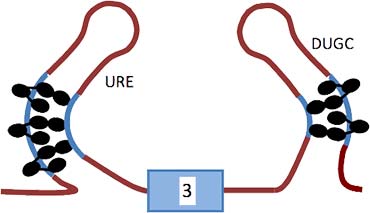
The diagram shows the model based on our results for the repression of exon 3 by PTB and associated proteins. Molecules of PTB are represented by ovals on the URE (upstream regulatory element) to the left and a UGC rich region (DUGC) to the right. Five or six molecules of PTB are associated with the transcript; three are associated through domains 1–3 with P3 and two or three similarly with DY. RRM4 of each PTB molecule is proposed to contact a relatively close C-rich tract.
Our aim now is to provide a complete description of the variety of assemblages forming on a pre-mRNA and to establish what defines an active complex. We are currently developing methods for monitoring the flexibility and dynamics of these complexes at the single molecule level, since we suspect that these overall properties of the complex integrate the inputs of the many individual bound proteins.
This work has connections to the development of therapies that exploit the vulnerability of splicing to nudges that push it to use alternative sites. We were involved in the first experiments to show that modified oligonucleotides could redirect splicing in the endogenous dystrophin gene. Our collaborators have pursued this, leading to the first approved drug for splice site switching, which targets certain cases of muscular dystrophy.
We have also developed a novel strategy for activating a specific splice, which is an important approach for therapy in spinal muscular atrophy. In the event, another splice-switching oligonucleotide was developed by others that proved to be more feasible; this has now been approved and is the first drug to halt the progression of the disease. In collaboration with Cyril Dominguez and Glenn Burley (Strathclyde), we are interested in the mechanisms by which four-stranded internal structures in the RNA (G4s) affect splicing and, in particular, switch splicing to produce pro-apoptotic isoforms of the genes Bcl-X and Mclk-1. By identifying molecules that bind to these structures and stabilise them, we hope to develop drugs that are of use in the treatment of some cancers.
Group members
Shijie Cui, Christian Lucas, Sumera Tubasum, Vasileios Pascalis, Max Wills, Philippe de Gusmao Araujo.
Publications
Selected publications:
- Jobbins, A.M., Campagne, S, Weinmeister, R., Lucas, C.M., Gosliga, A.R., Clery, A.,Chen, L., Eperon, L.P., Hodson, M.J., Hudson, A.J.H., Allain, F.H.T. and Eperon, I.C.* (2022) Exon-independent recruitment of SRSF1 is mediated by U1 snRNP stem-loop 3. EMBO J. 41: e107640.
- Bhogadia, M., Stone, B., Del Villar Guerra, R., Muskett, F.W., Ghosh, S., Taladriz-Sender, A., Burley, G.A., Eperon, I.C., Hudson, A.J., and Dominguez, C. (2022). Biophysical characterisation of the Bcl-x pre-mRNA and binding specificity of the ellipticine derivative GQC-05: implication for alternative splicing regulation. Frontiers in Molecular Biosciences, 9:943105. doi: 10.3389/fmolb.2022.943105.
- Bueno-Alejo, C.J., Santana Vega, M., Chaplin, A., Farrow, C., Axer, A., Burley, G.A., Dominguez, C., Kara, H., Paschalis, V., Tubasum, S., Eperon, I.C., Clark, A.W., and Hudson, A.J. (2022). Surface passivation with a perfluoroalkane brush improves the precision of single molecule measurements. ACS Appl. Mater. Interfaces 14, 49604−49616.
Supervision
Mechanisms of splice site selection/ single molecule microscopy/applications of chemical biology
Teaching
- BS3010: Gene expression: molecular basis and medical relevance
- BS2091: From genes to proteins
Press and media
RNA splicing